The idea of reverse engineering biology seems like a paradox. How could we ever reverse engineer anything for which we didn’t “invent” the original technology? Radios, bridges, computers—all these are human-made, built from logical, clean cut, organized systems. Biology, on the other hand, is wild and messy, complex in ways we can’t even conceive of. How could we reverse engineer biology—a millennia of evolution—when we don’t fully understand the complexities or processes of that system in the first place?
In fact, it’s for just those reasons that we must learn to reverse engineer biology. Otherwise, engineering new applications in biology will be limited to only those areas where we have a clean, logical, organized understanding of the biological system. To intervene in complex biological contexts we don’t fully understand but desperately need to affect—like disease, or aging, or cancer—reverse engineering may be our best tool. The future of bio will look much more like programming than pipetting. If we can unlock the power of reverse engineering in the field of biology, we may be able to break through all the supposed limits of engineering biology as we know it today.
To intervene in complex biological contexts we don’t fully understand but desperately need to affect—like disease, or aging, or cancer—reverse engineering may be our best tool.So what does that actually mean? To reverse engineer biology means applying the engineering concept of taking apart a process or mechanism in order to understand it and re-engineer it (perhaps in a new way)—and applying it to the biological world. That requires an intersection of two very different mindsets, which in some ways feel like fundamentally opposed approaches.
In his now classic piece (first published in 2002), Yuri Lazebnik posed the question of whether reverse engineering is possible in such a discovery-based science. Could a biologist fix a broken radio without instructions or knowledge of the radio? Probably not, he wrote; the approach that biology takes to understand biological systems—i.e., examining the effects of perturbations to suss out functional relationships—would give a biologist some sense of the parts and interactions, but not enough to begin to fit those parts together. Could a physicist fix that radio? Perhaps; physicists are trained to discover and understand the fundamental nature of physical laws and processes—but not necessarily the technology that comes from those laws of physics. So—could an electrical engineer fix a radio they had never seen before? Not right away; first, they would have to reverse engineer it—take apart the radio, examine it, identify the parts and how they work together. And then, yes. Using the knowledge gained through that approach, the radio could in all likelihood be reverse engineered, and broken processes fixed.
Despite the difference in approaches, biologists and engineers are actually both involved in the process of discovery, though in different ways.
In the first two cases, the very approach of the science itself seems at odds with what is needed to fix the radio. But despite the difference in approaches, biologists and engineers are actually both involved in the process of discovery, though in different ways. Biologists are trying to understand what evolution created and how; engineers are using their understanding in order to build something new.
So let’s go back to whether or not we can reverse engineer biology. How might an electrical engineer look at the problem? In a sense, this represents the foundation of synthetic biology, whether the goal is engineering proteins with new function and stability, or engineering cells with novel circuits and new function. To do so, we need certain key tools and concepts from both worlds. Below are some of those key concepts which bring both words together.
#1: Shared languages
As Lazebnik pointed out, one of the biggest failings of combining the approaches of discovery with engineering is the lack of a shared language. Lazebnik compared the language of biological circuits to that of electronic engineers:
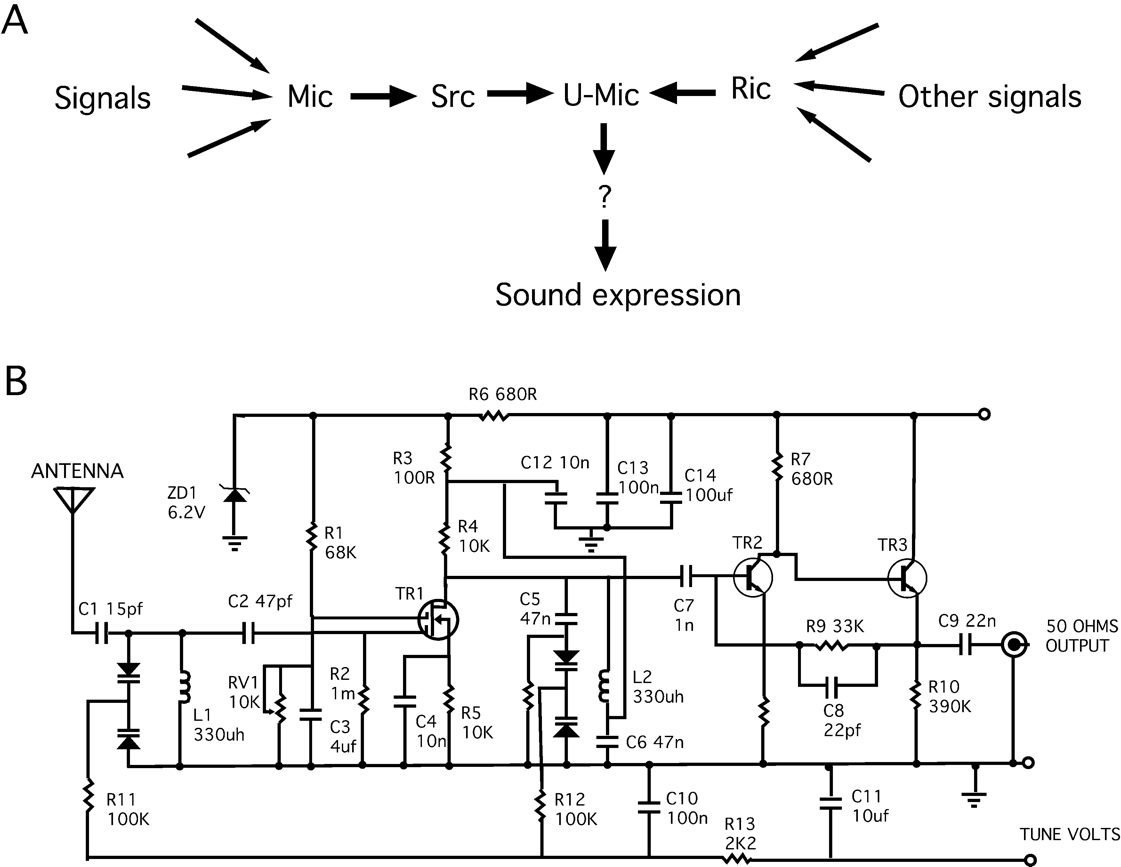
Figure A is the way a biologist would describe pathways, i.e. the “circuit” of interactions between proteins. If a biologist studied a radio, they would find interconnections between different parts, and map them out in broad pathways like the ones here. The circuit in Figure B is the way an engineer would represent these pathways — with much more quantitative and predictive properties, and much more detail on interactions represented.
Historically, the language biologists use has generally been less quantitative simply because the experiments themselves were not quantitative. But we are reading biological information on a quantitative level as we never have before, from genomics to proteomics to metabolomics to cell sorting.
So now, quantitative languages can now be deployed. Even simply the application of ordinary and partial differential equations brings a quantitative language not typically present before. But more sophisticated quantitative languages are beginning to be used as well: Verilog—a computer language used to engineer electronic circuits from radios to complex microprocessors—is already being used to model, predict, and even engineer biological genetic circuits.
# 2: Basic Building Blocks
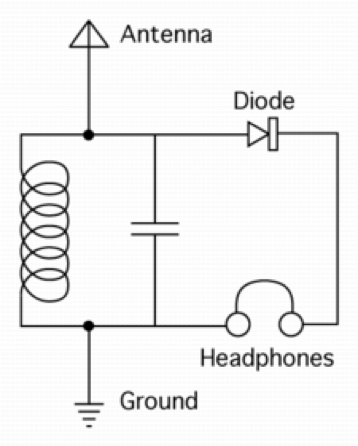
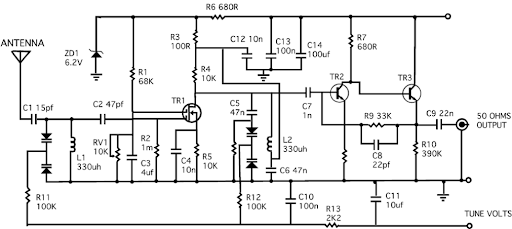
Different biological systems have different levels of complexity at different scales, from proteins to cells to tissues. One of the key principles of reverse engineering is to start with the simple system first—say, a simple radio circuit versus a complicated one (again, from Lazebnik):
(vs)
The simple radio circuit (top) would be considerably easier to reverse engineer—and still gives a fundamental understanding of the key components needed. The takeaway here is to start simple, and then add complexity. Not surprisingly, this very much has been at the core of how biology is studied, starting with simple organisms and then increasing in complexity (fruit flies to mice to humans).
# 3: Learn fast and iterate
Part of the challenge of biology tends to be the pace of experiments. To reverse engineer, you need to try something, test it, see if it works. Reverse engineering isn’t impossible if you get one test result a month (or a year)… but it would be very slow.
In order to design biology, we need to be able to probe with rapid cycles and different kinds of perturbations. New technologies in robotics and lab automation are already enabling both higher throughput, greater reproducibility, and faster results, even as quickly as in a single day. And the rise of chemical biology have given us the ability to engineer and apply new tool compounds—new molecules, proteins, and cells with which to perturb the system—with a diversity and scale previously not possible. Creating new tool compounds is one way to “tweak the parts” that make up a process, allowing you to both learn more about that process and to create new ones.
In engineering, you can “copy / paste / edit,” and build something better from there—instead of starting with a clean sheet of paper each time (as most therapeutics were previously developed). CAR T therapy—i.e, the engineering of T-cells for therapeutic purposes—is another example like this, where each successive generation of CAR T therapeutics builds upon the design of the previous one.
# 4: Swap out parts
If using building blocks like legos and “tweaking the parts” are some ways to understand or change a biological system, “part swapping” is yet another important concept and tool. In this context, “part swapping” would mean a well-defined change where you leverage all the biology of the known parts in order to swap things around and learn more.
Imagine trying to reverse engineer a lamp, and swapping out light bulb to learn what works and what doesn’t. Gene editing tools like CRISPR are natural instruments for reverse bioengineering: we can literally swap out components into living organisms—even multiple parts at once, and systemically—and then measure the changes. Today we are just beginning at the gene level, but one day will be able to go to greater scales: replacing numerous genes at once to alter large, multi-protein complexes; maybe even radically altering cells to swap to different cell phenotypes.
When bio becomes programming…
Combining these two different worlds requires an intersection and translation of very different worlds and cultures—but doing so has incredible potential and power. If we can reverse engineer biology, we can begin to design—and program—biology itself.
References:
[1] https://www.cell.com/cancer-cell/fulltext/S1535-6108(02)00133-2
[2] https://blog.udemy.com/reverse-engineering-tutorial/
[3] https://en.wikipedia.org/wiki/Reverse_engineering
[4] http://legacy.lincolninteractive.org/html/CES%20Introduction%20to%20Engineering/Unit%203/u3l7.html
http://legacy.lincolninteractive.org/samples/introduction-to-engineering