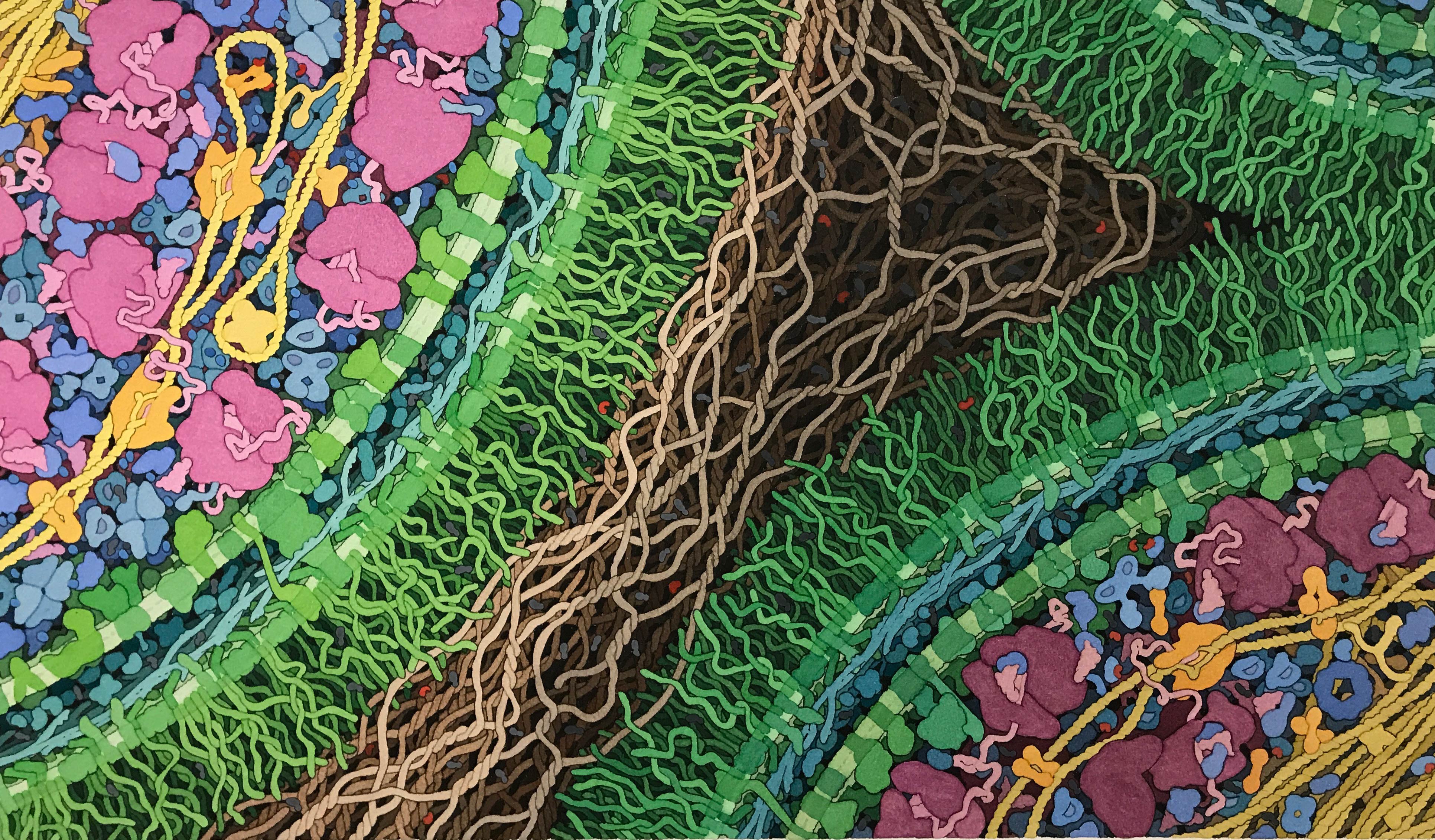
cells harboring an engineered genetic circuit by David Goodsell for Asimov
Modern computer chips are a marvel of human engineering. With billions of transistors, they are among the most complex devices we’ve ever created, yet they operate with precision and digital accuracy. How did this become possible? Certainly not by a person laying down each transistor by hand. Instead, a software pipeline and ecosystem of tools — built around electronic design automation (EDA) — gives people superpowers to build things that we couldn’t otherwise do.
With software, we can design and program a whole world of applications on top of chips and computers. When it comes to engineering biology this way though, we’re way behind. Despite the fact that cells are the original computers — processing information, encoding computational operations, communicating with each other, organizing spatially, and so on — we’re still at the very beginning of biological circuit design, even though the ideas have been around for decades. Biological circuits are sets of biochemical reactions that have similar components (logic gates, memory, etc.) as computer circuits, but that are driven by the chemistry of life — with molecules taking the role of electrons and electricity.
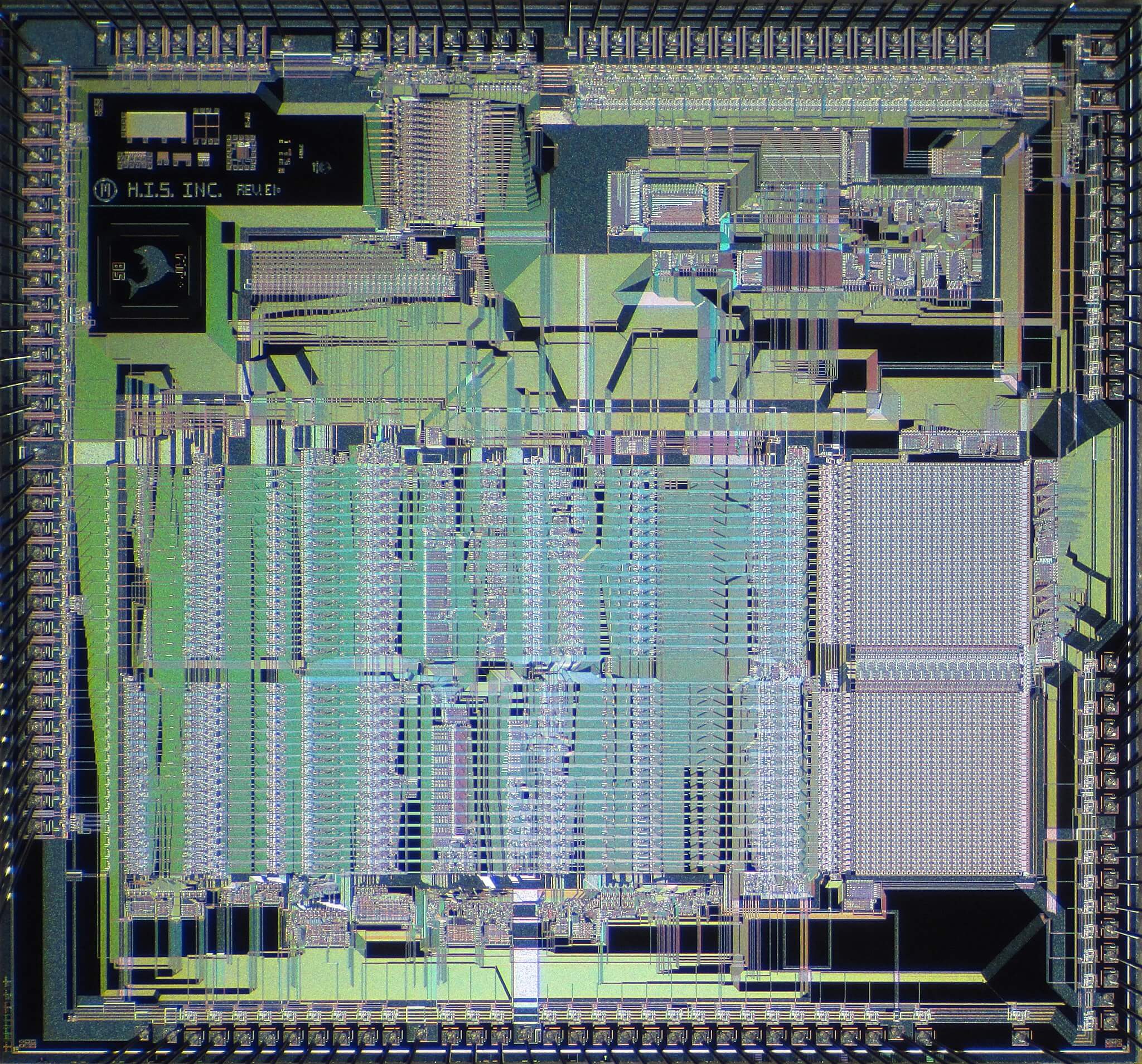
microprocessor circa 1987; with engineering economics (Moore’s Law) today’s microprocessors are incredibly more complex [image: Pauli Rautakorpi]
This approach allows the same tools and mindsets learned from software to be applied to the biological realm. Take hierarchy, a key aspect to software design: One does not design every transistor in a modern microprocessor by hand, but instead designs it in modular parts (e.g. circuits to do memory, arithmetic, logic, control, etc.) that are then combined. With hierarchy, you don’t need to understand the inner workings of those parts; they are abstracted away, allowing people to design high-level goals without needing to get stuck in the details. In biological circuit design, a modular approach that decouples the genetic context and interactions allows bioengineers to build more reliable systems. It also allows recent software engineering practices — such as DRY (don’t repeat yourself) and agile programming — to play an important role in biological engineering.
But even if we can design complex biological circuits this way, the question is… would they work?
The next step — in both the electronic and biological context — is to predict the outcomes of circuits, because making new chips as well as new cells is expensive at the prototype stage. EDA tools include powerful simulators of circuits, so engineers can debug them virtually, resulting in a low-cost, high-turnaround process. Asimov’s custom tools include a powerful simulator that can predict whether or not a biological circuit will work with up to 90% accuracy. This is a huge improvement over doing it with the physical device where readouts are more limited… Or where thousands to potentially millions of designs must be brute-force tested to find the one that works — as is the case when you only have empiricism, and not engineering, on your side.
The hallmark of Asimov’s ability to engineer chips is that it avoids a slow, expensive, empirical trial-and-error approach.
Not only do such biological circuit design automation tools give bioengineers the ability to debug biological circuits much like we debug software — with complete detail of what the simulated circuit is doing — but Asimov engineers have also developed modular biological circuit components that don’t have adverse reactions to other parts of the cell. Why does this matter? It’s akin to a computer programmer designing code that is then injected into a running program or existing operating system. These biological building blocks can be easily used downstream by circuit designers — the bio advance in turn facilitates the computer science advance, namely the accurate simulation of biological circuits.
With Asimov’s approach, high-accuracy simulation, and circuit building-blocks, we can greatly speed the development of biological circuits — decreasing their cost, and greatly increasing their sophistication and complexity. Continuing with our analogy of computers here, we’re still in the “transistor phase” of things, so are not yet at the point where the full complexity of a modern microprocessor can be realized into the circuits of cells. But there are many initial applications where this technology can make major advances — much like how early microprocessors, as simple as they were, became a dramatically enabling technology.
Because biology is everywhere, living cells have applications in everything from food and materials to agriculture to healthcare. In fact, 7 of the top 10 drugs today are biologics, i.e., proteins that have therapeutic properties. These proteins are manufactured in cells at the cost of billions of dollars. Asimov’s technology could drive a dramatic reduction in cost to patients — enabling these drugs to be in the hands of more and more people.
Looking even further out, a bolder application for designing biological circuits is one where new cellular therapies could sense disease in the body, perform logic, and drive a precise curative response — like therapeutic, microscopic “bio-robots”. Sounds like science fiction, but what EDA brought us for electronic circuits could also have once been considered science fiction: It helped bring the reality today of massive cloud supercomputers and smartphones that would make even Captain Kirk jealous.
To help realize the vision of bringing this biological-circuits tech platform to broader applications, I’m excited to announce Andreessen Horowitz’ investment in Asimov, led by co-founders Alec Nielsen, Raja Srinivas, Chris Voigt, and Doug Densmore. Alec, Chris, and Doug co-authored the seminal paper, “Genetic Circuit Design Automation” in Science describing the initial technology. Alec and Raja met while doing their PhDs in Biological Engineering at MIT, and Raja brings both a computational biology background along with previous entrepreneurial experience. Chris is a professor of Biological Engineering at MIT where he leads the Voigt Lab in pioneering synthetic biology research, and Doug is a professor of Electrical and Computer Engineering at Boston University where he directs a cross-disciplinary group on design automation for engineering biology. With this combination of both scientific and computer science expertise cutting across disciplines, they’re the best team to bring this technology to fruition… and I’m honored to join the Asimov board.
image credits: photo of an original painting by David Goodsell for Asimov; Bull DPS 6000 CPU die by Pauli Rautakorpi via Wikimedia Commons (CC3.0)



